Welcome to BloodChIP Xtra
Introduction
Hematopoiesis is one of the best characterised developmental systems in vertebrates, serving as a model system for the multi-lineage differentiation of adult stem cells. Hematopoietic stem cells (HSCs) maintain production of circulating blood cells via their capacity to either self-renew or differentiate to mature cell types. The most primitive HSCs have multilineage potential but give rise to progenitor cells with increasing lineage restriction (1,2). However, the components and hierarchy of the gene regulatory networks which control identity of human adult HSCs and their progeny, and the mechanisms by which these are hijacked or reactivated in leukemic cells, remain incompletely understood.
Lineage determining transcription factors (TFs) and the genomic regions they bind and regulate are a key component of GRNs that control cell identity (3,4). A core set of seven TFs (heptad: ERG, FLI1, GATA2, RUNX1, TAL1, LYL1, and LMO2), all known regulators of healthy and leukemic haematopoiesis, work in combination to regulate gene expression in hematopoietic stem and progenitor cells (HSPCs) (5-7). Heptad TFs also function in the context of other genome regulators including histone modifications and formation of chromatin loops. Knowledge of how TFs cooperate with each other and interact with other genome regulators to regulate gene expression during HSPC differentiation is key to improving our understanding of normal blood development and how these processes are corrupted in leukaemia.
What is BloodChIP Xtra?
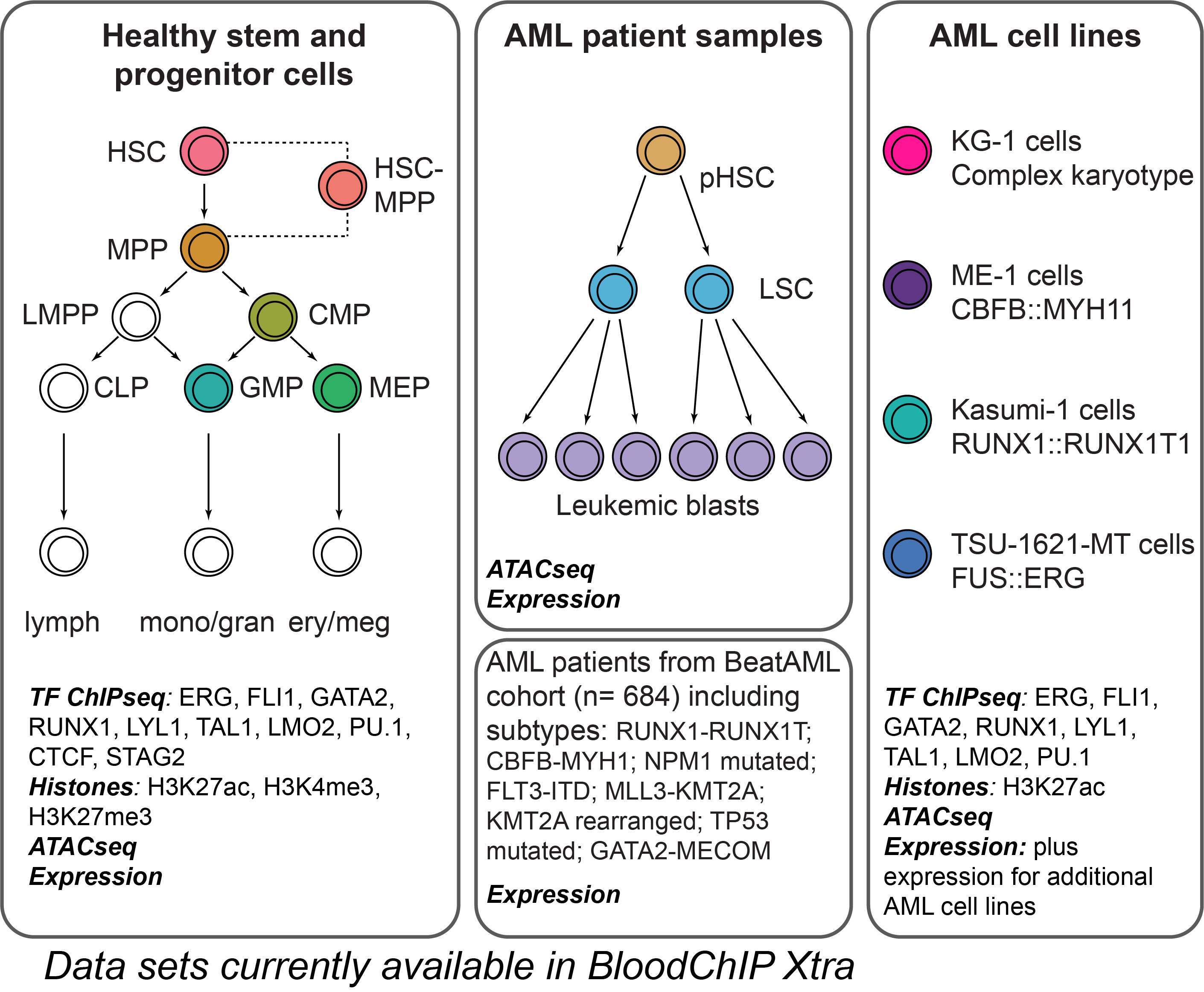
BloodChIP Xtra is a user-friendly database that facilitates genome-wide exploration and visualization of transcription factor (TF) occupancy and chromatin configuration in rare primary human hematopoietic stem (HSC-MPP) and progenitor (CMP, GMP, MEP) cells and acute myeloid leukemia (AML) cell lines (KG-1, ME-1, Kasumi1, TSU-1621-MT), along with chromatin accessibility and gene expression data from these and primary patient AMLs. This database features significantly more datasets than our earlier offering BloodChIP (8). An interactive web interface allows users to explore the regulatory landscape of any gene across multiple blood cell types, and then associate the epigenetic/regulatory profile with gene expression across healthy and leukemic cells. All queries, as well as the complete database, can be exported by the user for further data analysis.
To start using BloodChIP Xtra click here
How to cite
Julie A I Thoms, Forrest C Koch, Alireza Raei, Shruthi Subramanian, Jason W H Wong, Fatemeh Vafaee, John E Pimanda (2023) BloodChIP Xtra: an expanded database of comparative genome-wide transcription factor binding and gene-expression profiles in healthy human stem/progenitor subsets and leukemic cells. Nucleic Acids Research, https://doi.org/10.1093/nar/gkad918
References
- Doulatov, S., Notta, F., Laurenti, E. and Dick, J.E. (2012) Hematopoiesis: a human perspective. Cell Stem Cell, 10, 120-136.
- Laurenti, E. and Gottgens, B. (2018) From haematopoietic stem cells to complex differentiation landscapes. Nature, 553, 418-426.
- Pimanda, J.E. and Gottgens, B. (2010) Gene regulatory networks governing haematopoietic stem cell development and identity. Int J Dev Biol, 54, 1201-1211.
- Thoms, J.A.I., Beck, D. and Pimanda, J.E. (2019) Transcriptional networks in acute myeloid leukemia. Genes Chromosomes Cancer, 58, 859-874.
- Beck, D., Thoms, J.A., Perera, D., Schutte, J., Unnikrishnan, A., Knezevic, K., Kinston, S.J., Wilson, N.K., O'Brien, T.A., Gottgens, B. et al. (2013) Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood, 122, e12-22.
- Subramanian, S., Thoms, J.A.I., Huang, Y., Cornejo Paramo, C.P., Koch, F.C., Jacquelin, S., Shen, S., Song, E., Joshi, S., Brownlee, C.P. et al. (2023) Genome-wide Transcription Factor binding maps reveal cell-specific changes in the regulatory architecture of human HSPC. Blood.
- Thoms, J.A.I., Truong, P., Subramanian, S., Knezevic, K., Harvey, G., Huang, Y., Seneviratne, J.A., Carter, D.R., Joshi, S., Skhinas, J. et al. (2021) Disruption of a GATA2-TAL1-ERG regulatory circuit promotes erythroid transition in healthy and leukemic stem cells. Blood, 138, 1441-1455.
- Chacon, D., Beck, D., Perera, D., Wong, J.W. and Pimanda, J.E. (2014) BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res, 42, D172-177.