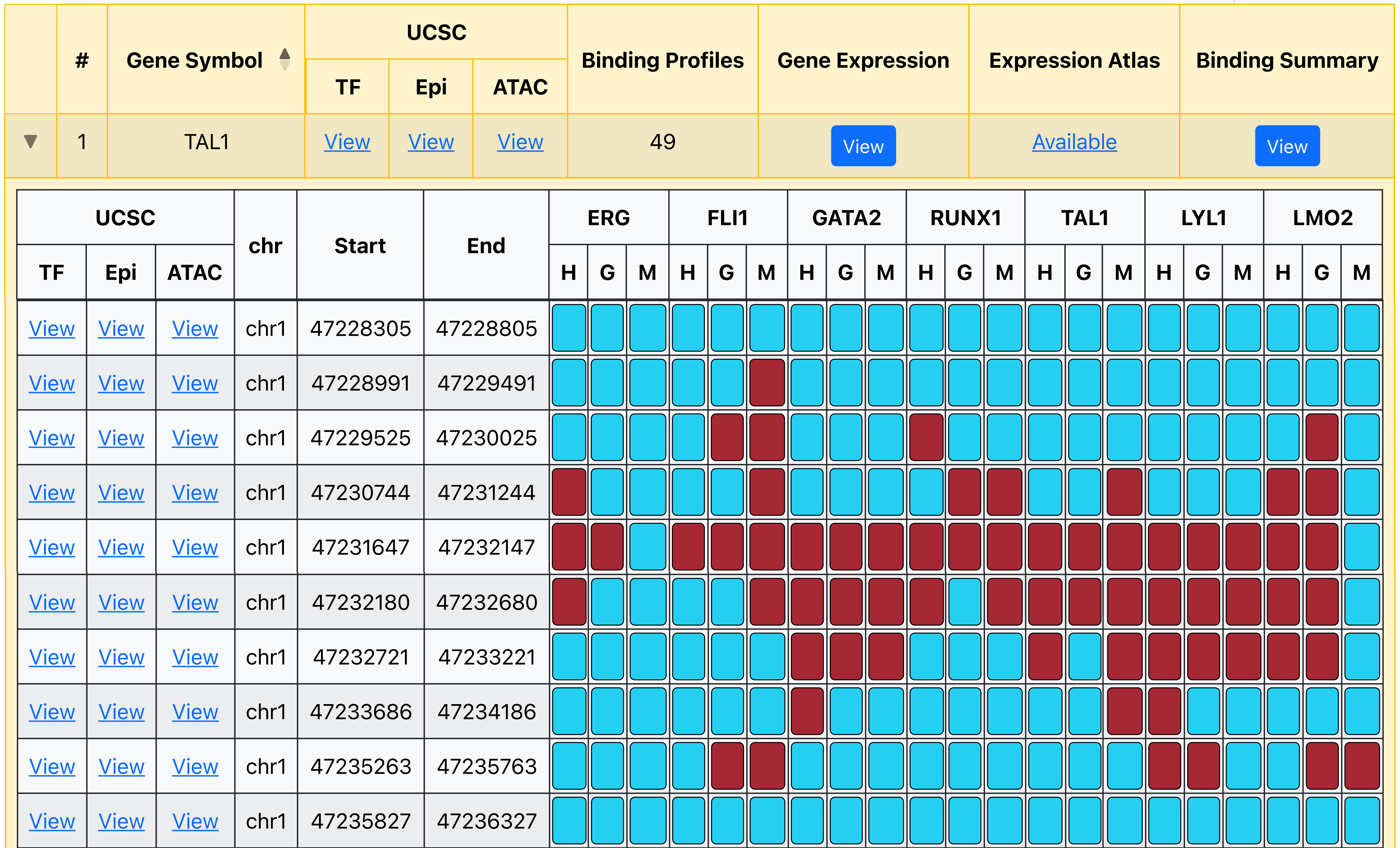
Searching by gene
To retrieve transcription factor binding profiles for specific gene(s), a list of gene symbols (i.e. HHEX, ETV6, etc.) can be entered one per line. Genes can also be specified using wild cards, for example "ETV*" would retrieve all genes beginning with "ETV" (i.e. ETV1, ETV2, etc.). The figure below illustrates a search using a list of 5 genes.
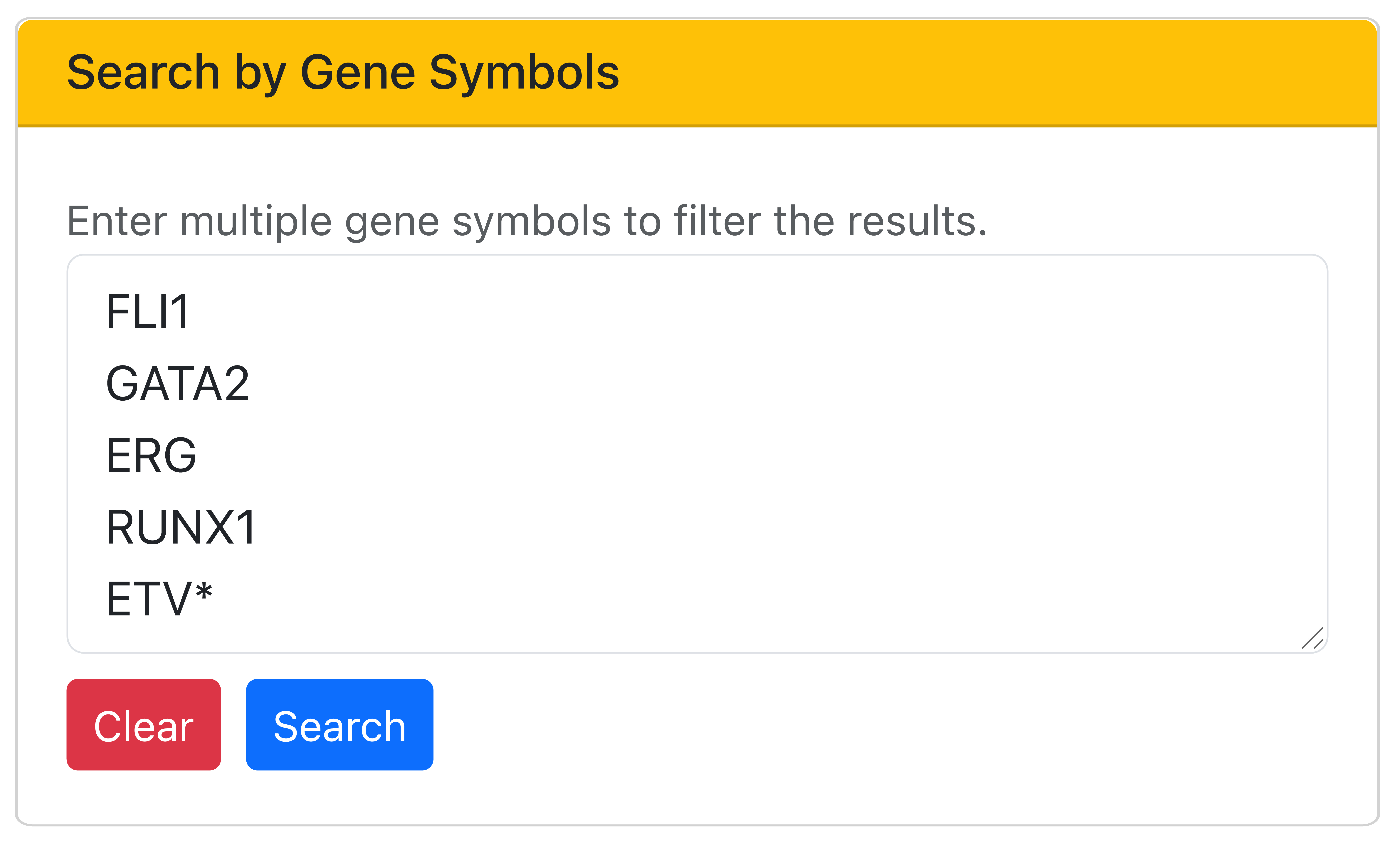
Searching by transcription factor binding
BloodChIP Xtra also allows users to retrieve all genes either bound by a combination of selected TFs or by any of the selected TFs in the desired cell type. To retrieve genes that are regulated by multiple TFs across multiple cell types, the "AND" condition can be selected between each of the selected TFs. To retrieve all genes bound by any of the selected TFs, the "OR" condition can be selected. The example figure below illustrates how to retrieve all genes combinatorially bound by ERG, FLI1, GATA2, RUNX1, LYL1, TAL1, and LMO2 in HSCs.
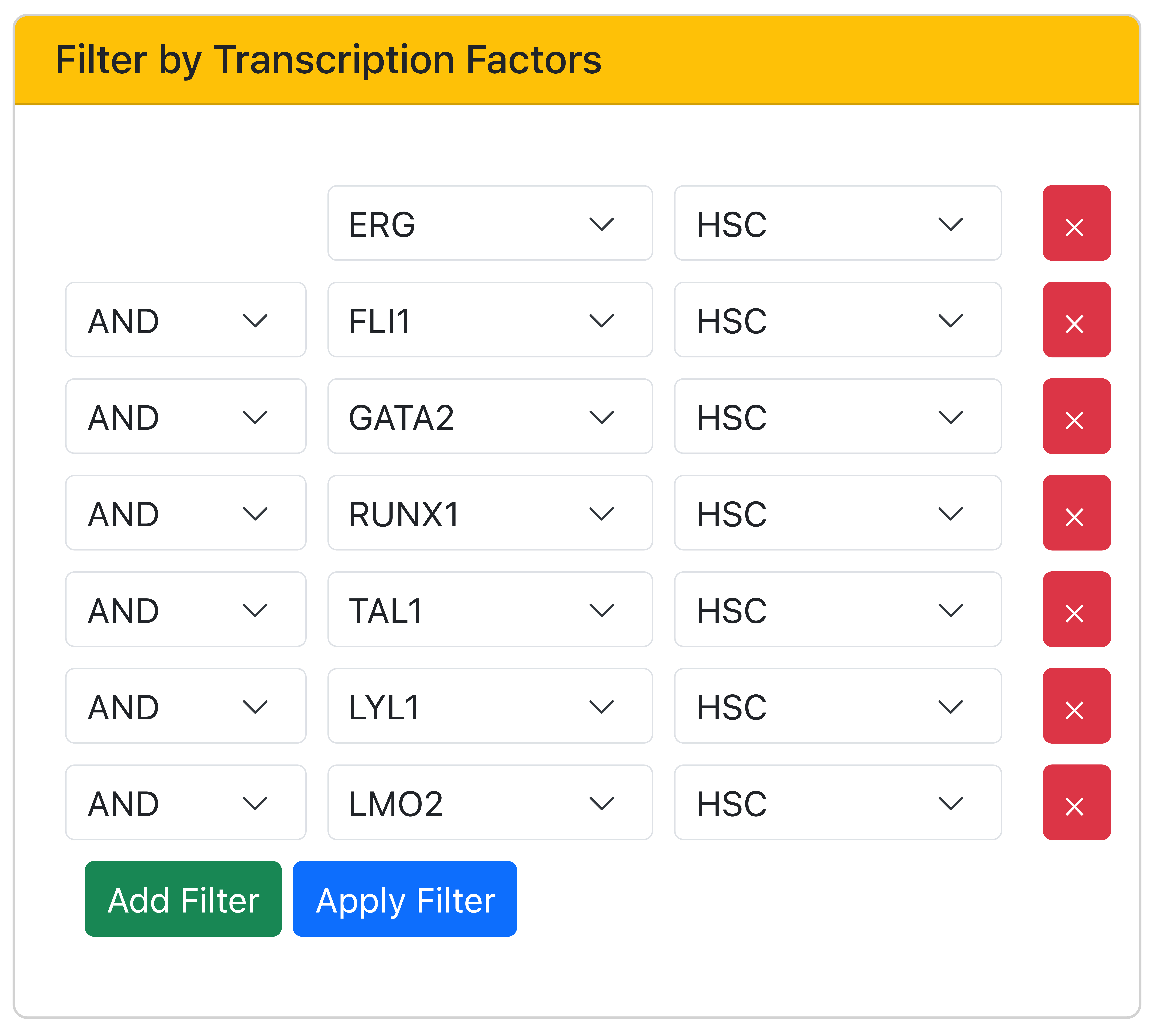
The Gene Database table
The genes retrieved by a query are displayed in a table underneath the results. Each gene entry can be expanded to show TF peaks associated with the gene. In each TF peak row, the binding status of selected factor (TF, histone, ATAC) in each cell type is shown. A red box indicates that the particular factor in the particular cell type is binding at this peak. A blue box indicates that there is no binding and a grey box indicates that the TF binding profile is unavailable for the particular cell type. Apart from displaying TF binding information, the table also provide links to UCSC tracks for the visualisation of TF binding profiles, Epigenetic modifiers, and ATACseq at a specific gene or peak locus. Furthermore, for each gene, a link is provided to a downloadable graph showing expression of that gene across multiple cell types, a summary table indicating whether any element belonging to that gene is bound by any factor, and also to the Expression Atlas.